Research
Visualizing Escherichia coli sub-cellular structure using sparse deconvolution spatial light interference tomography
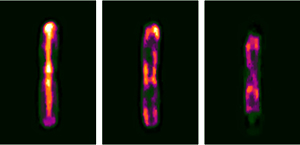
M. Mir*, S. D. Babacan*, M. Bednarz, M. N. Do, I. Golding, and G. Popescu, PloS ONE, 2012.
*Equal contribution
Abstract: Studying the 3D sub-cellular structure of living cells is essential to our understanding of biological function. However, tomographic imaging of live cells is challenging mainly because they are transparent, i.e., weakly scattering structures. Therefore, this type of imaging has been implemented largely using fluorescence techniques. While confocal fluorescence imaging is a common approach to achieve sectioning, it requires fluorescence probes that are often harmful to the living specimen. On the other hand, by using the intrinsic contrast of the structures it is possible to study living cells in a non-invasive manner. One method that provides high-resolution quantitative information about nanoscale structures is a broadband interferometric technique known as Spatial Light Interference Microscopy (SLIM). In addition to rendering quantitative phase information, when combined with a high numerical aperture objective, SLIM also provides excellent depth sectioning capabilities. However, like in all linear optical systems, SLIM’s resolution is limited by diffraction. Here we present a novel 3D field deconvolution algorithm that exploits the sparsity of phase images and renders images with resolution beyond the diffraction limit. We employ this label-free method, called deconvolution Spatial Light Interference Tomography (dSLIT), to visualize coiled sub-cellular structures in E. coli cells which are most likely the cytoskeletal MreB protein and the division site regulating MinCDE proteins. Previously these structures have only been observed using specialized strains and plasmids and fluorescence techniques. Our results indicate that dSLIT can be employed to study such structures in a practical and non-invasive manner.
Sparse Bayesian Methods for Low-Rank Matrix Estimation

S. D. Babacan, M. Luessi, R. Molina, and A. K. Katsaggelos, IEEE Transactions on Signal Processing, 2012.
Abstract: Recovery of low-rank matrices has recently seen significant activity in many areas of science and engineering, motivated by recent theoretical results for exact reconstruction guarantees and interesting practical applications. In this paper, we present novel recovery algorithms for estimating low-rank matrices in matrix completion and robust principal component analysis based on sparse Bayesian learning (SBL) principles. Starting from a matrix factorization formulation and enforcing the low-rank constraint in the estimates as a sparsity constraint, we develop an approach that is very effective in determining the correct rank while providing high recovery performance. We provide connections with existing methods in other similar problems and empirical results and comparisons with current state-of-the-art methods that illustrate the effectiveness of this approach.
Cell imaging beyond the diffraction limit using sparse deconvolution spatial light interference microscopy (dSLIM)
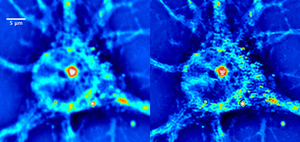
S. D. Babacan, Z. Wang, M. N. Do, and G. Popescu, Biomedical Optics Express, vol. 2 no. 7, pp. 1815-1827, 2011.
(one of top downloaded papers (>1,000 downloads) in 2011 in Biomedical Optics Express)Abstract: We present an imaging method, dSLIM, that combines a novel deconvolution algorithm with spatial light interference microscopy (SLIM), to achieve 2.3x resolution enhancement with respect to the diffraction limit. By exploiting the sparsity properties of the phase images, which is prominent in many biological imaging applications, and modeling of the image formation via complex fields, the very fine structures can be recovered which were blurred by the optics. With experiments on SLIM images, we demonstrate that significant improvements in spatial resolution can be obtained by the proposed approach. Moreover, the resolution improvement leads to higher accuracy in monitoring dynamic activity over time. Experiments with primary brain cells, i.e. neurons and glial cells, reveal new subdiffraction structures and motions. This new information can be used for studying vesicle transport in neurons, which may shed light on dynamic cell functioning. Finally, the method is flexible to incorporate a wide range of image models for different applications and can be utilized for all imaging modalities acquiring complex field images.
Paper Link | ECI Presentation | QLI Lab website for more information on SLIM
Reference-Guided Sparsifying Transform Design for Compressive Sensing MRI
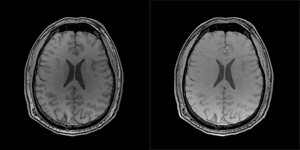
S. D. Babacan, X. Peng, X.-P. Wang, M. N. Do, Z.-P. Liang, Engineering in Medicine and Biology Conference (EMBC), Boston, July, 2011.
Abstract: Compressive sensing (CS) MRI aims to accurately reconstruct images from undersampled k-space data. Most CS methods employ analytical sparsifying transforms such as total-variation and wavelets to model the unknown image and constrain the solution space during reconstruction. Recently, nonparametric dictionary-based methods for CS-MRI reconstruction have shown significant improvements over the classical methods. These existing techniques focus on learning the representation basis for the unknown image for a synthesis- based reconstruction. In this paper, we present a new frame- work for analysis-based reconstruction, where the sparsifying transform is learnt from a reference image to capture the anatomical structure of unknown image, and is used to guide the reconstruction process. We demonstrate with experimental data the high performance of the proposed approach over traditional methods.
Variational Bayesian PCA
- S. Nakajima, M. Sugiyama, S. D. Babacan, "On Bayesian PCA: Automatic Dimensionality Selection and Analytic Solution," in International Conference on Machine Learning (ICML 2011), Bellevue, Washington, June 2011.
- S. Nakajima, M. Sugiyama, S. D. Babacan, "Global Solution of Fully-Observed Variational Bayesian Matrix Factorization is Column-Wise Independent," in Neural Information Processing Systems (NIPS 2011), Granada, Spain, December 2011
Summary: In probabilistic PCA, the fully Bayesian estimation is computationally intractable. To cope with this problem, several types of approximation schemes were introduced. In our ICML 2011 paper, it is shown that a simplified variant of variational Bayesian PCA (VB-PCA), where the latent variables and loading vectors are assumed to be mutually independent, is the most advantageous in practice because its empirical Bayes solution experimentally works as well as the original VB-PCA, and its global optimal solution can be computed efficiently in a closed form. In NIPS 2011, we have shown that the global solution of VB-PCA is column-wise independent, leading to diagonal covariances. This shows that the global solution obtained using the simplified variant is also the optimal solution of VB-PCA.
ICML 2011 Paper | NIPS 2011 Paper | NIPS 2011 poster slides | Nakajima's website for more information and papers on variational Bayesian PCA
Bayesian symmetrical EEG/fMRI fusion with spatially adaptive priors
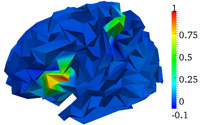
M. Luessi, S. D. Babacan, R. Molina, J.R. Booth, and A. K. Katsaggelos, NeuroImage, vol. 55, issue 1, pp. 113-132, March 2011.
Abstract: In this paper, we propose a symmetrical EEG/fMRI fusion method which combines EEG and fMRI by means of a common generative model. The use of a total variation (TV) prior as well as spatially adaptive temporal priors allows for adaptation to the local characteristics of the estimated responses. We utilize a fully Bayesian formulation with a variational Bayesian framework and obtain a fully automatic fusion algorithm. Simulations with synthetic data and experiments with real data from a multimodal study on face perception demonstrate the performance of the proposed method.